By Vikas Kumar, Yan Wang, and Lawrence Rasouliyan | OMNY Health
Extracting Meaningful Insights from Free-Text EHR Data
Extracting meaningful information from unstructured data is never easy, It’s often a time-consuming task. In structured EHR data for instance, certain values such as diagnosis codes are often insufficient to capture the full context. The same is mostly true for gastrointestinal bleeding, where a slight inaccuracy might cause significant implications for both diagnosis and treatment.
One such challenge which our team at OMNY Health tried to solve is the differentiation between two forms of GI bleeding: melena, black tarry stools, usually caused by upper GI bleeding, and hematochezia, bright red blood in stool, usually from lower GI bleeding. Although both are captured under general ICD-10 codes, namely K92.1 and K92.2, respectively, these do not make a distinction between the two. Our objective was to bridge this gap using unstructured data and machine learning.
Building the Model: From Clinical Notes to Meaningful Insights
We conducted a retrospective observational study using the OMNY Health Real-World Data Platform (2017–2025). Patients with ICD-10 codes starting with “K” (gastrointestinal diseases) or “E” (endocrine diseases) were included. A clinical domain expert reviewed 1,000 random clinical notes from patients with GI bleed–related codes (K92.1 or K92.2) to identify phrases that indicated either melena or hematochezia.
Through this manual review, our team identified 28 phrases for melena and 51 phrases for hematochezia, which were then used to build two separate N-gram models. These models searched millions of notes across the dataset to identify encounters associated with each condition.
These models are validated against real-world clinical outcomes to ensure reliability. We compared the rates of upper versus lower GI diagnoses, endoscopic procedures (EGD vs. colonoscopy), and pharmacologic treatments within 30 days following each encounter.
Results: Real-World Validation That Reflects Clinical Reality
Our validation showed that the N-gram models accurately differentiated between the two GI bleeding types.
- Precision: 96% for melena; 98% for hematochezia
- Recall: 7.9% for melena; 5.3% for hematochezia
TABLE 1 — Samples of Phrases Used for Melena and Hematochezia N-gram
(Note: samples shown; complete list of phrases used to train each model is available in OMNY Health’s internal dataset.)
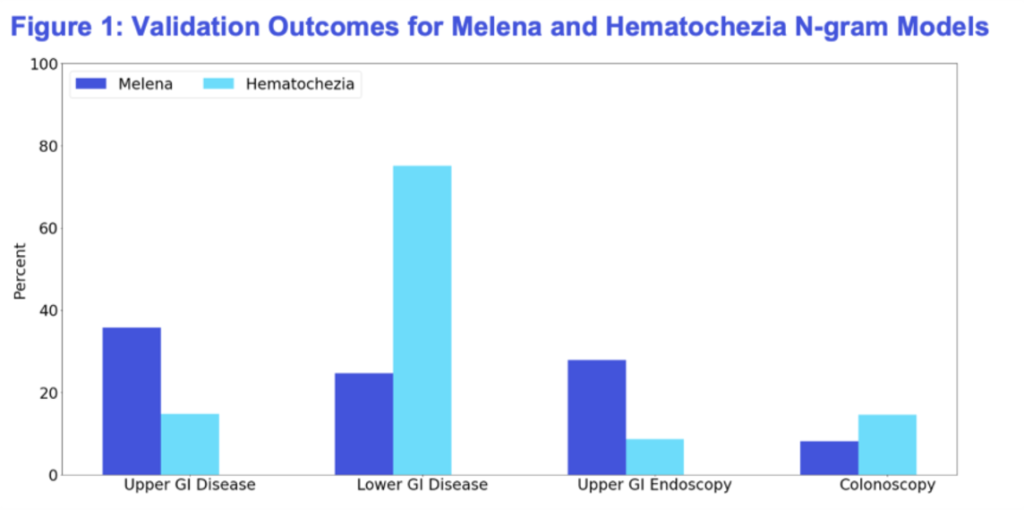
Patients identified with melena were more likely to have an upper GI diagnosis and to undergo esophagogastroduodenoscopy (EGD). Conversely, patients with hematochezia were more likely to have lower GI diagnoses and receive colonoscopy procedures. These results aligned closely with clinical expectations, reinforcing the accuracy and validity of our models.
Figure 1. Validation Outcomes for Melena and Hematochezia N-gram Models
Why It Matters: A Step Toward Richer Real-World Evidence
The ability to differentiate between melena and hematochezia in unstructured EHR data proves to be more beneficial for more granular, clinically meaningful insights. This allows researchers and healthcare organizations to:
- Better characterize patient populations
- Refine outcome measures for GI bleeding studies
- Support drug safety and effectiveness research with higher precision
OMNY Health platform helps researchers to unlock the full potential of real-world clinical information, i.e. turning free-text notes into actionable insights, hence improving care delivery and research quality.
Looking Ahead
The study demonstrates how natural language processing (NLP) can be effective in bridging gaps in structured EHR data. As we continue validating these models, our primary focus remains on empowering researchers, clinicians, and life science partners with trustworthy, real-world data solutions.
© 2025 OMNY Health
